Genome editing with CRISPR-Cas9
The Alt-R™ CRISPR-Cas9 System includes all the reagents needed for successful genome editing in your research applications based on the natural S. pyogenes CRISPR-Cas9 system.
Point, click, edit. Guaranteed.*
Ordering
- Benefit from the latest improvements with on- and off-target design and chemical modifications
- Enjoy easy ordering of custom or predesigned guide RNAs
- Get desirable editing results with high on-target potency and reduced off-target activity with Alt-R HiFi CRISPR-Cas9 nuclease
- Expect precision control editing with efficient delivery of the RNP by lipofection or electroporation
- Protect cells from toxicity or innate immune response activation
CRISPR-Cas9 gRNA
Guide RNAs (gRNAs) contain the target-specific sequence for guiding Cas9 protein to a genomic location. We offer 3 gRNA formats: crRNA:tracrRNA duplex, crRNA XT:tracrRNA duplex, and single guide RNA (sgRNA).
CRISPR-Cas9 crRNA
To be used with tracrRNA to form a functional gRNA duplex. Suitable for most applications. Contains chemical modifications to protect from degradation by cellular RNases.
CRISPR-Cas9 crRNA XT
To be used with tracrRNA to form a functional gRNA duplex. Suitable for challenging experimental conditions (e.g., high nuclease environments or with Cas9 mRNA). Contains additional chemical modifications compared to crRNA to provide a cost-effective option for increased functional stability.
CRISPR-Cas9 sgRNA
Single RNA molecules comprised of both crRNA and tracrRNA sequences. Suitable for challenging experimental conditions (e.g., high nuclease environments or with Cas9 mRNA). Contains chemical modifications for a high level of functional stability.
Guaranteed editing with predesigned gRNA designs
We guarantee* our predesigned guide RNAs targeting human or mouse, rat, zebrafish, or nematode genes. For other species, we recommend using our proprietary algorithms that enable the design of custom guide RNAs.
Note: If you have protospacer designs of your own, or designs from publications, use our design checker tool to assess their on- and off-targeting potential before ordering guide RNAs that are synthesized with using our Alt-R gRNA modifications.
Use your own designs
If you have protospacer designs of your own, or designs from publications, we recommend using the design checker tool to assess on- and off-targeting potential before ordering guide RNAs that are synthesized with our Alt-R guide RNA gRNA modifications. For designs that do not require this analysis, you can directly order any user-defined crRNA or sgRNA with our tube and plate ordering buttons.
CRISPR cGMP gRNA Manufacturing Service
Are you looking to accelerate your CRISPR genome editing program from discovery to clinical trials? Our Engineering Run and full cGMP guide RNA manufacturing services offer a streamlined and regulatory-compliant solution. Backed by comprehensive documentation, our guide RNAs simplify regulatory filings, offering a straightforward path to clinical success. Click here to learn more.
Alt-R HiFi Cas9 Nuclease with improved selectivity
The new Alt-R HiFi Cas9 Nuclease has similar on-target potency to wild-type S.p. Cas9, but with significantly reduced off-target effects, allowing for precise genome editing, even under challenging conditions.
GMP CRISPR Nucleases
Does your project require high-quality cGMP nucleases? SpCas9 Nuclease and SpyFi® Cas9 Nuclease are available for immediate delivery at cGMP quality. Learn how IDT can help you rapidly transition from the lab to clinical trials. Contact us today.
Custom CRISPR solutions
For more flexibility in guide RNA ordering, CRISPR Custom Guide RNAs allow a wide range of lengths and modifications to support a variety of CRISPR systems and experiments.
Don’t see what you’re looking for?
Have questions for our CRISPR experts? Your time is valuable and we’ll prioritize your inquiry. Click on “Request a consultation” to provide brief information about your project, and we’ll be in touch to discuss it ASAP.
Request a consultation* We guarantee that predesigned Alt-R CRISPR-Cas9 guide RNAs will provide successful editing at the target site, when delivered as a ribonucleoprotein complex as described in the Alt-R Protocols, using Alt-R CRISPR-Cas9 guide RNAs (crRNA:tracrRNA duplex or sgRNA) and either Alt-R S.p. Cas9 nuclease or Alt-R S.p. HiFi Cas9 nuclease. Analysis of editing must be at the DNA level, such as with the Alt-R Genome Editing Detection Kit or DNA sequencing. If successful editing is not observed for a predesigned guide RNA while an appropriate positive control is successful; a one-time “no-cost” replacement of the predesigned Alt-R CRISPR-Cas9 guide RNA will be approved, upon contacting us. This guarantee does not extend to any replacement product, or to any other incurred or incidental costs or expenses.
Product Details
What makes the Alt-R CRISPR-Cas9 System so great?
- Exact length determination of the crRNAs for increased activity
- Chemical modifications to help increase nuclease resistance and reduce innate immune responses
- Knowledge that our native Cas9 and HiFi Cas9 variant has shown increased selectivity in our studies
- Two-part guide RNA (crRNA:tracrRNA) provides high activity at an affordable price
- Unique supporting reagents (electroporation enhancer, labeled tracrRNA, controls) provide successful editing
- Complete collection of user resources, including guides, protocols, and demonstrated protocols
Alt-R CRISPR-Cas9 System
Simple delivery of ribonucleoprotein complexes (crRNA:tracrRNA:Cas9 or sgRNA:Cas9).
Figure 1. Overview of Alt-R CRISPR-Cas9 System experiments for ribonucleoprotein delivery by lipid-mediated transfection, electroporation, or microinjection using (A) 2-part guide RNAs (crRNA:tracrRNA duplex) or (B) sgRNAs.
CRISPR-Cas9 genome editing methods use a Cas9 endonuclease to generate double-stranded breaks in DNA. Cas9 endonuclease requires a CRISPR RNA (crRNA) to specify the DNA target sequence, and the crRNA must be combined with the transactivating crRNA (tracrRNA) to activate the endonuclease and create a functional editing ribonucleoprotein complex (Figure 1A). In an alternative approach, the crRNA and tracrRNA can be delivered as a single RNA oligonucleotide (Figure 1B). After cleavage, DNA is then repaired by non-homologous end-joining (NHEJ) or homology-directed recombination (HDR), resulting in a modified sequence. Alt-R CRISPR-Cas9 reagents and kits provide essential, optimized tools needed to use this pathway for genome editing research.
| Alt-R CRISPR-Cas9 System | |
|---|---|
| Ribonucleoprotein components | |
Option 1: Alt-R CRISPR-Cas9 crRNA:tracrRNA | Alt-R CRISPR-Cas9 crRNA
|
Alt-R CRISPR-Cas9 tracrRNA
| |
| Option 2: Alt-R CRISPR-Cas9 sgRNA |
|
| Alt-R S.p. Cas9 Nuclease/Nickase |
|
| Additional reagents and kits | |
|---|---|
| Alt-R CRISPR-Cas9 Control Kits |
|
| Alt-R CRISPR-Cas9 Electroporation Enhancer | For primary and difficult-to-transfect cells |
| Alt-R HDR Enhancer V2 | For improved rates of homology-directed repair |
| Alt-R Genome Editing Detection Kit | For mutation identification and estimating editing efficiency |
Ribonucleoprotein components
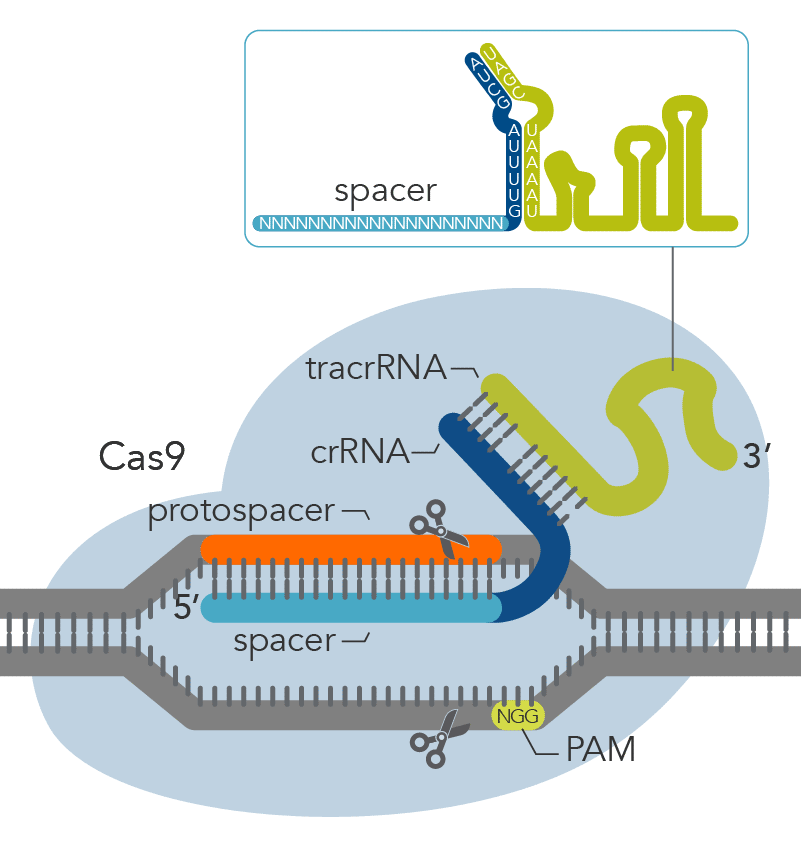
The Alt-R CRISPR-Cas9 System offers two options for generating synthetic guide RNAs. The two-part system pairs an optimized, shortened universal tracrRNA oligonucleotide (67 nt) with an optimized, shortened, target-specific crRNA oligonucleotide (36 nt) for improved targeting of Cas9 to dsDNA targets (Figure 2). The single guide RNA (sgRNA) option combines the crRNA and tracrRNA segments into one long RNA molecule, reducing the number of components and simplifying the CRISPR workflow.
Figure 2. Components of the Alt-R CRISPR-Cas9 System for directing Cas9 endonuclease to genomic targets. For the 2-part guide RNA format, the crRNA:tracrRNA complex uses optimized Alt-R crRNA and tracrRNA sequences that hybridize and then form a complex with Cas9 endonuclease to guide targeted cleavage of genomic DNA. Alternatively, the sgRNA (not shown), which contains relevant crRNA and tracrRNA sequences, forms a complex with Cas9 endonuclease to guide targeted cleavage of genomic DNA. The cleavage site is specified by the spacer element of the crRNA (light blue bar). The crRNA spacer element recognizes 19 or 20 nt on the strand opposite from the NGG PAM site. The PAM site must be present immediately downstream of the protospacer element for cleavage to occur. Research by IDT scientists has shown that the Alt-R CRISPR-Cas9 System provides the highest percentage of on-target genome editing when compared to competing designs of native S. pyogenes crRNA:tracrRNA.
While delivering Cas9 nuclease as part of an RNP is the preferred method, the Alt-R CRISPR-Cas9 System is also compatible with S. pyogenes Cas9 from any source, including cells that stably express S. pyogenes Cas9 endonuclease, or when Cas9 is introduced as a DNA or mRNA construct.
Option 1 (2-part guide RNA)
Alt-R CRISPR-Cas9 crRNA
All Alt-R CRISPR-Cas9 crRNAs are 35–36 nt RNA oligos containing the 19 or 20 nt target-specific protospacer region, along with the 16 nt tracrRNA fusion domain. We recommend 20 nt protospacers for most applications. crRNAs should be duplexed with Alt-R CRISPR-Cas9 tracrRNA before RNP complex formation.
Alt-R CRISPR-Cas9 crRNAs are synthesized with proprietary chemical modifications, which protect the crRNA from degradation by cellular RNases and further improve on-target editing performance. When using 2-part gRNAs under highly challenging conditions (e.g., high nuclease environments or with Cas9 mRNA), use Alt-R CRISPR-Cas9 crRNA XT, which have additional chemical modifications for the highest level of stability and function.
We guarantee* our predesigned guide RNAs targeting human, mouse, rat, zebrafish, or nematode genes. For other species, we recommend using our proprietary algorithms to design custom guide RNAs. If you have protospacer designs of your own or from publications, use our design checker tool to assess their on- and off-targeting potential before ordering guide RNAs that are synthesized using our Alt-R guide RNA modifications.
* See Ordering section for details.
Alt-R CRISPR-Cas9 tracrRNA
The 67 nt Alt-R tracrRNA is much shorter than the classical 89 bases of the natural S. pyogenes tracrRNA. We find that shortening the tracrRNA increases on-target performance. Alt-R CRISPR tracrRNA also contains proprietary chemical modifications that confer increased nuclease resistance.
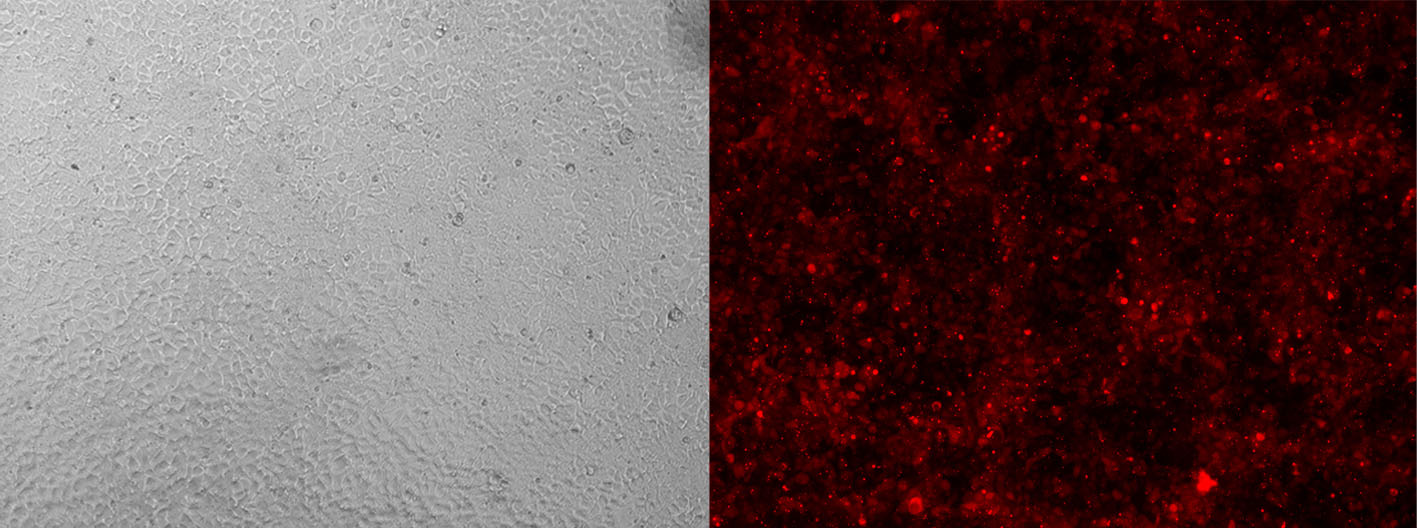
Alt-R CRISPR-Cas9 tracrRNA labeled with ATTO™ 550 (ATTO-TEC) provide the same function as their unlabeled counterparts. However, the fluorescent dye allows you to monitor transfection or electroporation efficiency during preliminary experiments to optimize transfection conditions in your cell types (Figure 3).
Figure 3. Detection of fluorescently labeled tracrRNA by fluorescence microscopy. HEK-293 cells stably expressing Cas9 nuclease were reverse transfected (RNAiMAX reagent, Thermo Fisher Scientific) with Alt-R CRISPR-Cas9 crRNA (unlabeled) complexed with Alt-R CRISPR-Cas9 tracrRNA – ATTO 550 (final concentration of 10 nM). Images were taken 48 hr after transfection. Magnification: 10X.
Labeled tracrRNAs can also help concentrate transfected cells via FACS (fluorescence-activated cell sorting) analysis, which can simplify your screening process for cells with CRISPR events.
Alt-R CRISPR tracrRNA orders include Nuclease-Free Duplex Buffer for forming the complex between crRNA and tracrRNA oligos. Alt-R tracrRNA can be ordered in larger scale and paired with all of your target specific crRNAs, allowing for an easy and a cost-effective means of studying many CRISPR sites.
Option 2 (single guide RNA)
Alt-R CRISPR-Cas sgRNA
Alt-R CRISPR-Cas9 sgRNAs are long RNA oligonucleotides (99–100 bases) containing the target-specific crRNA region and the Cas9-interacting tracrRNA region within a single molecule (i.e., 19–20 base protospacer region and 80-base universal sgRNA region). Like other Alt-R RNAs, it contains chemical modifications to stabilize the RNA, increasing resistance to nuclease activity. For challenging conditions (e.g., high nuclease environments or with Cas9 mRNA), sgRNAs may provide increased potency.
Cas9 endonucleases
Alt-R S.p Cas9 nuclease
The Alt-R S.p. Cas9 Nuclease V3 enzyme is a high purity, recombinant S. pyogenes Cas9. The enzymes include nuclear localization sequences (NLSs) and C-terminal 6-His tags. The S. pyogenes Cas9 enzyme must be combined with a gRNA to produce a functional, target-specific editing complex. For the best editing, combine the Alt-R S.p. Cas9 Nuclease V3 enzyme with the optimized Alt-R CRISPR gRNA in equimolar amounts.
Alt-R S.p. HiFi Cas9 nuclease
The Alt-R S.p. HiFi Cas9 Nuclease V3 offers improved specificity over wild-type Cas9, greatly reducing the risk of off-target cutting events. This Cas9 variant also preserves the high level of editing efficiency expected from a Cas9 nuclease, maintaining 90–100% on-target editing activity at most sites. For applications that are sensitive to off-target events, combining the Alt-R S.p. HiFi Cas9 Nuclease V3 with optimized Alt-R CRISPR-Cas9 gRNA (crRNA:tracrRNA) is highly recommended.
Alt-R S.p. Cas9-GFP (or RFP) nuclease
The Alt-R S.p. Cas9-GFP V3 and S.p. Cas9-RFP V3 nucleases are high purity, recombinant S. pyogenes Cas9 enzymes that are expressed as fusion proteins with nuclear localization sequences (NLSs) and C-terminal 6-His tags. These enzymes have on-target functionality comparable to wild-type S.p. Cas9 and are designed for applications that require post-transfection visualization of the protein or enrichment of edited cells using fluorescence-activated cell sorting (FACS). These enzymes should be combined with Alt-R CRISPR gRNA in equimolar amounts.
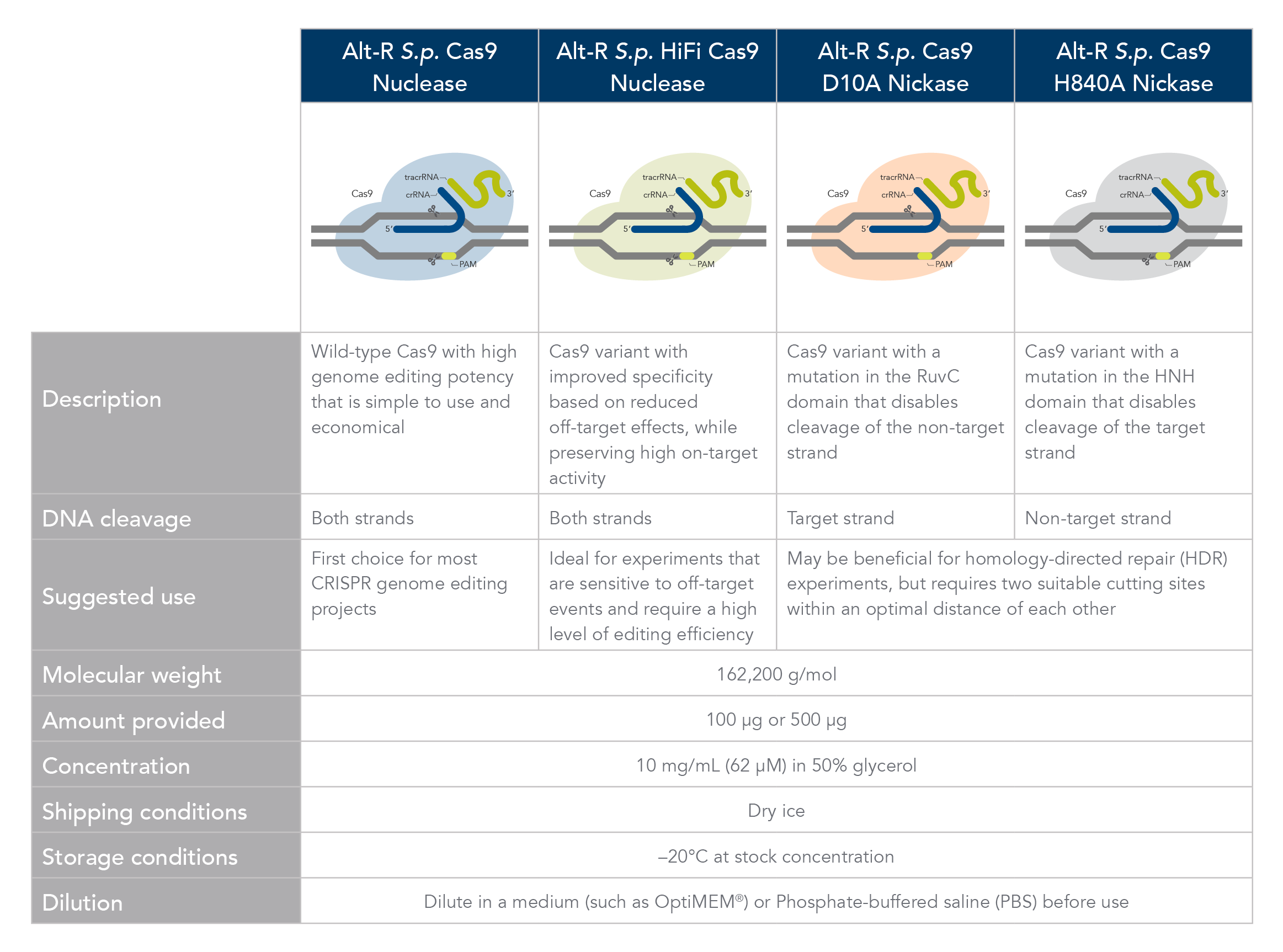
Alt-R S.p. Cas9 nickases
Cas9 nickases allow specific cutting of only one strand at the DNA target site. Cuts to both strands of DNA are accomplished by using either Alt-R S.p. Cas9 D10A Nickase V3 or Alt-R S.p. Cas9 H840A Nickase V3, with 2 gRNAs that target two neighboring Cas9 sites, one on either strand of the target region. This functionally increases the length of the recognition sequence from 20 to 40 bases. For more information about using Cas9 nickases, see this application note.
Alt-R S.p. dCas9 protein
Alt-R S.p. dCas9 Protein V3 has mutations that result in the loss of nuclease activity. This protein can form RNP complexes with Alt-R gRNAs and bind to the target region specified by the gRNA without cutting the DNA.
Like the other Alt-R enzymes, Alt-R S.p. dCas9 Protein V3 is provided as 10 mg/mL in 50% glycerol, and it can be diluted in PBS or Opti-MEM® media (Thermo Fisher) before use.
Cas9 comparison chart
Homology-directed repair reagents
Alt-R HDR Donor Oligos
Alt-R HDR Donor Oligos have been developed specifically for insertion into DNA by HDR. These HDR templates have enhanced stability and higher rates of incorporation than standard oligonucleotides.
- Exceptional flexibility: multiple species, as well as single or multiple (batch) design modes
- Compatible with Cas9 nucleases (double-stranded cleavage) or nickases (single-stranded cleavage)
- Accommodates incorporation of desired sequence (such as sequence tags, stop codons, GFP, enhancers, or promoters) into target site
Alt-R HDR Enhancer V2
Alt-R HDR Enhancer is a small molecule compound that increases homology-directed repair. Alt-R HDR Enhancer V2 is a new and improved small molecule compound that exhibits its activity in multiple cell lines, including both adherent and suspension cell lines. Its activity is independent of the enzyme employed; for example, it can be used either with Alt-R S.p. Cas9 nucleases or A.s. Cas12a (Cpf1) nucleases. This versatile reagent is also compatible with electroporation and lipofection methods.
Additional reagents and kits
Alt-R CRISPR-Cas9 Control crRNAs and PCR Assays
Optional controls for human, mouse, and rat are available for the 2-part Alt-R CRISPR-Cas9 System.
We recommend using the appropriate Alt-R CRISPR-Cas9 Control Kit for studies in human or mouse cells. The control kits include an Alt-R CRISPR HPRT Positive Control crRNA targeting the HPRT (hypoxanthine phosphoribosyltransferase) gene and a computationally confirmed Alt-R CRISPR-Cas9 Negative Control crRNA. The kit also includes the Alt-R CRISPR-Cas9 tracrRNA for complexing with the crRNA controls, Nuclease-Free Duplex Buffer, and validated PCR primers for amplifying the targeted HPRT region in the selected organism. The inclusion of the PCR assay makes these kits ideal for confirmation of HPRT modification using the Alt-R Genome Editing Detection Kit.
Alt-R control kit components can be ordered individually.
For information about sgRNA controls, contact us.
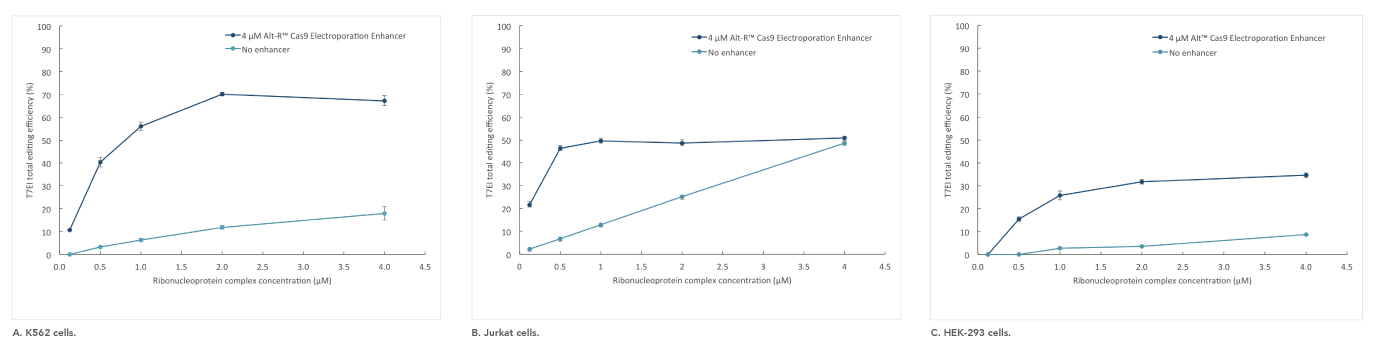
Alt-R Cas9 Electroporation Enhancer
If you are studying primary or hard-to-transfect cells, electroporation is often a viable alternative to lipid-based transfection in CRISPR experiments. The Alt-R Cas9 Electroporation Enhancer is a Cas9-specific carrier DNA that is optimized to work with the Amaxa® Nucleofector® device (Lonza) and Neon® System (Thermo Fisher) to increase transfection efficiency and thereby increase genome editing efficiency (Figure 4).
Figure 4. Alt-R Cas9 Electroporation Enhancer improves CRISPR editing efficiency in ribonucleoprotein (RNP) electroporation experiments. K562 (A), Jurkat (B), and HEK-293 (C) cells were transfected (Amaxa System, Lonza) with 0.125–4 µM RNP (Alt-R S.p. Nuclease 3NLS complexed with Alt-R CRISPR-Cas9 crRNA and tracrRNA). Electroporation reactions were performed in the presence (dark blue) or absence (light blue) of 4 µM Alt-R Cas9 Electroporation Enhancer.
Alt-R Genome Editing Detection Kit
Use this kit to identify on-target genome editing and estimate genome editing efficiency in CRISPR experiments. Learn more.
Product Data
Homology-directed repair (HDR)
Alt-R HDR Enhancer V2 improves HDR efficiency several fold
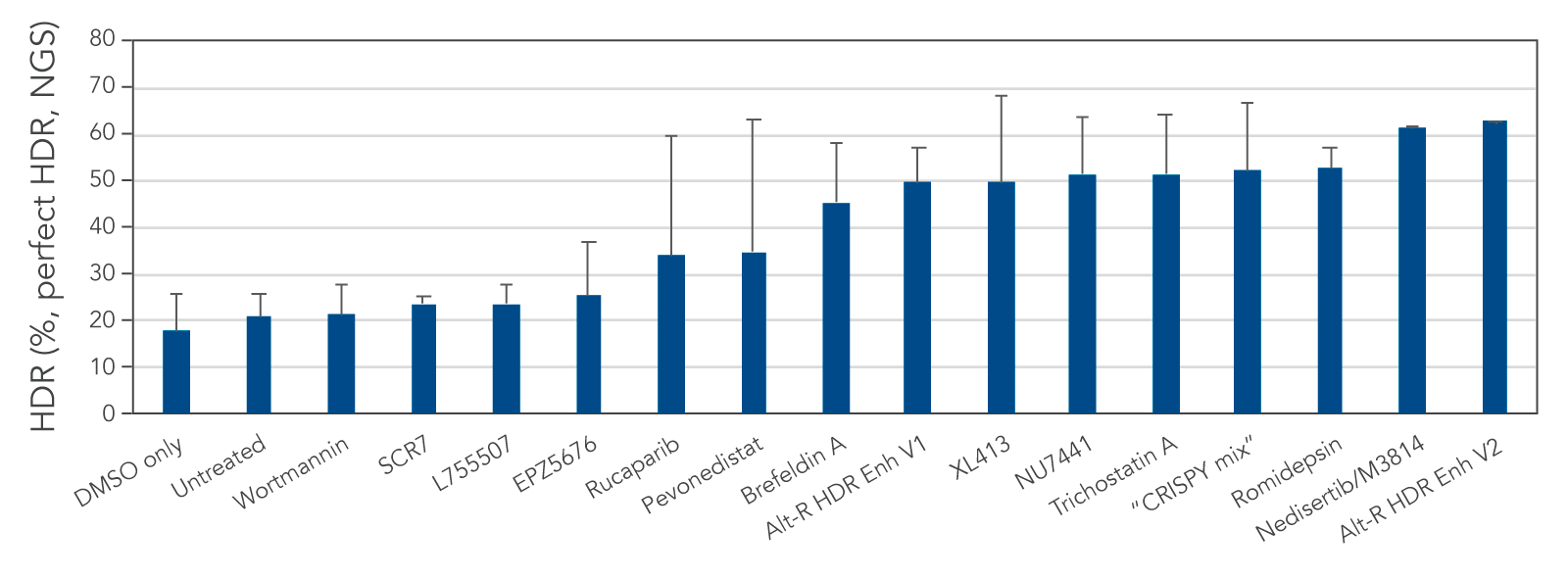
We tested several possible chemical compounds for their ability to improve HDR efficiency and found that our Alt-R HDR Enhancer V2 greatly increased the rate of HDR (Figure 1).
Figure 1. Alt-R HDR Enhancer V2 outperforms other small molecules from the literature. RNP complex (2 μM) targeting human HPRT1 and 3 μM Alt-R HDR Donor designed to insert an EcoRI restriction site were delivered together into Jurkat cells by electroporation using the 4D-Nucleofector™ System (Lonza). The RNP complex comprised Alt-R S.p. Cas9 Nuclease V3 complexed with Alt-R CRISPR-Cas9 crRNA and tracrRNA. Immediately after electroporation, cells were cultured in media containing no treatment, DMSO (vehicle control), or a recommended dose of small molecule (0.02 μM wortmannin, 5 μM SCR7, 5 μM L755507, 0.1 μM EPZ5676, 5 μM rucaparib, 1 μM pevonedistat, 0.1 μM brefeldin A, 30 μM Alt-R HDR Enhancer V1, 10 μM XL413, 2.5 μM NU7441, 10 μM trichostatin A, “CRISPY mix” [30 μM NU7026, 10 μM trichostatin A, 1 μM MLN4924], 0.1 μM romidepsin, 20 μM nedisertib/M3814, or 1 μM Alt-R HDR Enhancer V2). Genomic DNA was isolated 48 hours after lipofection, and target regions were amplified by PCR and sequenced on an Illumina™ MiSeq™ instrument. HDR efficiency was quantified using the IDT in-house NGS CRISPAltRations™ pipeline. The highest HDR rate was achieved when using the Alt-R HDR Enhancer V2 at 1 μM.
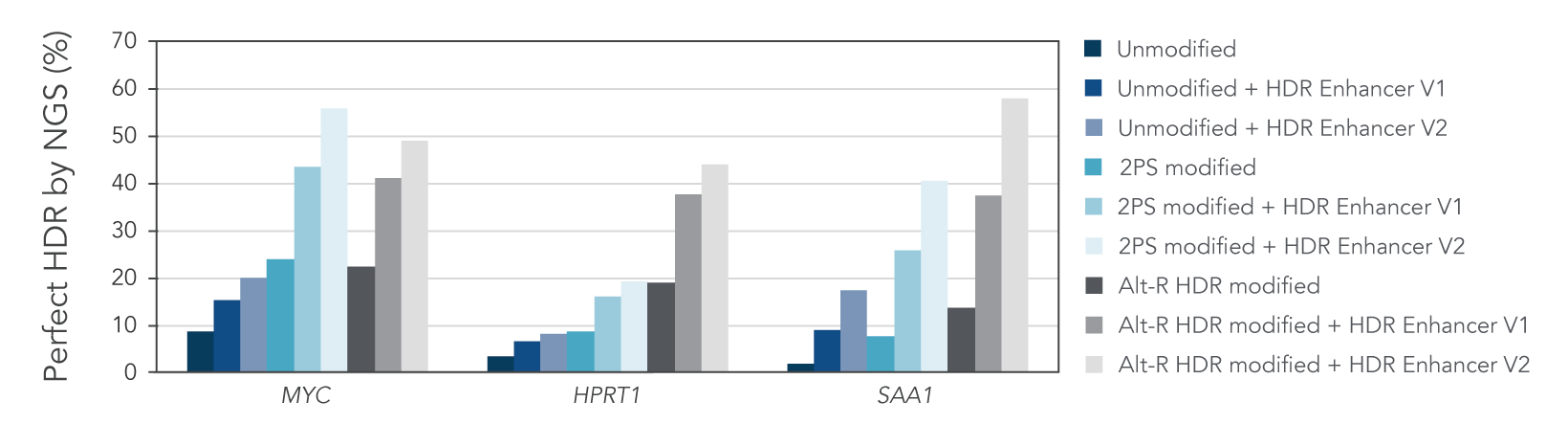
Alt-R HDR modified donors and Alt-R HDR Enhancer V2 lead to the higher HDR rates
We investigated whether combining Alt-R HDR Enhancer with Alt-R modifications of HDR donor oligos would further improve HDR rates. Our data showed that this works well, leading to higher HDR rates (Figure 2).
Figure 2. Alt-R HDR modified donors and Alt-R HDR Enhancer have an additive effect on HDR improvement. RNP complexes (2 μM) targeting three genomic loci (MYC, HPRT, and SAA1) along with 0.5 μM single-stranded Alt-R HDR donor oligo were delivered to HeLa cells by electroporation with 3 μM Alt-R Cas9 Electroporation Enhancer using the 4D-Nucleofector™ System (Lonza). The RNP complex comprised Alt-R S.p. HiFi Cas9 Nuclease V3 complexed with Alt-R CRISPR-Cas9 crRNA and tracrRNA. Unmodified, PS modified, or Alt-R HDR modified donor templates were used. Immediately after electroporation, cells were plated in media with either no treatment, 30 μM Alt-R HDR Enhancer (V1), or 1 μM HDR Enhancer V2. Genomic DNA was isolated 48 hours after electroporation, and HDR efficiency was measured by amplicon sequencing on an Illumina™ MiSeq™ system (v2 chemistry, 150 bp paired-end reads). The highest HDR rate was achieved with the combination of the Alt-R HDR Donor and HDR Enhancer V2.
Alt-R HDR Enhancer also improves HDR efficiency in cells transfected via lipofection (please see supplemental data).
Guide RNAs
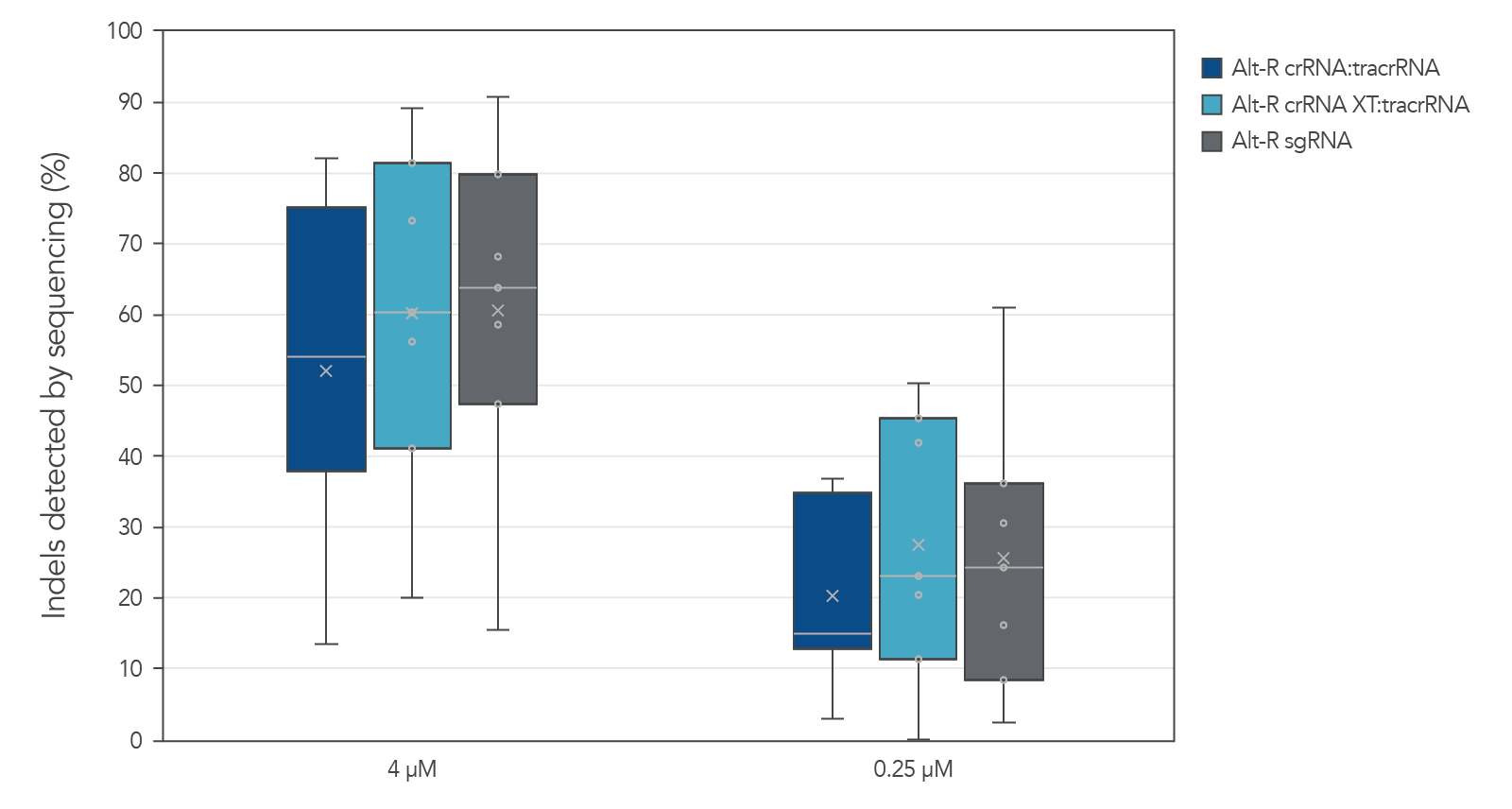
Alt-R guide RNA variants generate similar editing rates when used as part of ribonucleoprotein (RNP) complexes
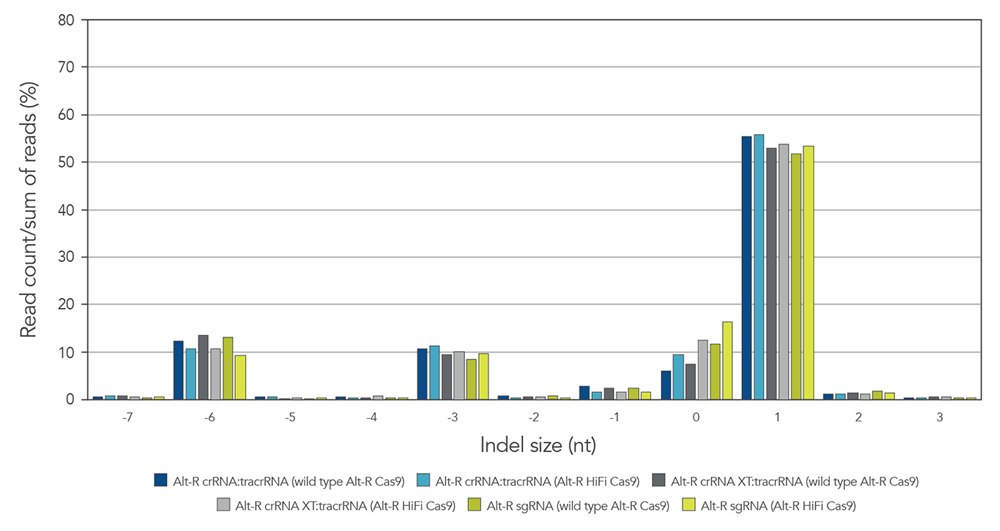
Guide RNAs (gRNAs) targeting 12 sites in the HPRT gene were delivered as RNP. Genome editing using 3 gRNA formats (crRNA:tracrRNA duplex, crRNA XT:tracrRNA duplex, and sgRNA) at two doses were examined. The comparable results are shown in a box-and-whiskers plot (Figure 3).
Figure 3. Similar genome editing by gRNA variants when delivered as RNP complexes. gRNA variants targeting 12 HPRT sites were
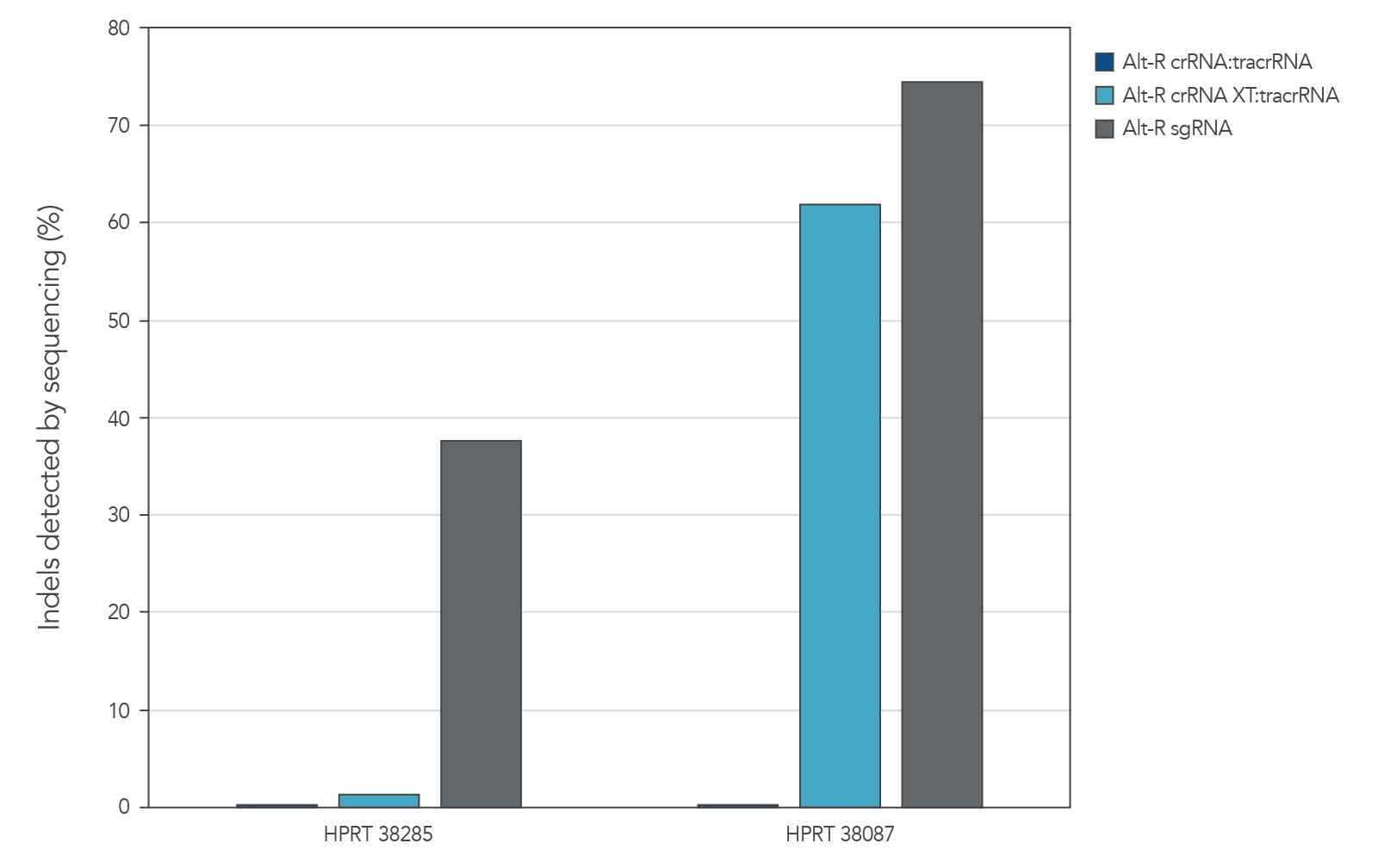
When co-delivering Cas9 mRNA with gRNA, the functional stability of the Alt-R guide RNAs becomes important for targeted genome editing
Genome editing activity of 3 gRNA formats (crRNA:tracrRNA duplex, crRNA XT:tracrRNA duplex, and sgRNA) was compared. The gRNA was co-delivered with Cas9 mRNA into cultured cells. The results show that at two HPRT sites, gRNA format can affect the rate of genome editing (Figure 4).
Figure 4. At some genome editing sites,
Alt-R guide RNA variants generate identical on-target repair profiles when used as part of RNP complexes
Guide RNAs (gRNAs) targeting the EMX1 gene were delivered as RNP that included either
Figure 5. gRNA variants produce identical on-target repair profiles. gRNA complexes targeting EMX1 were
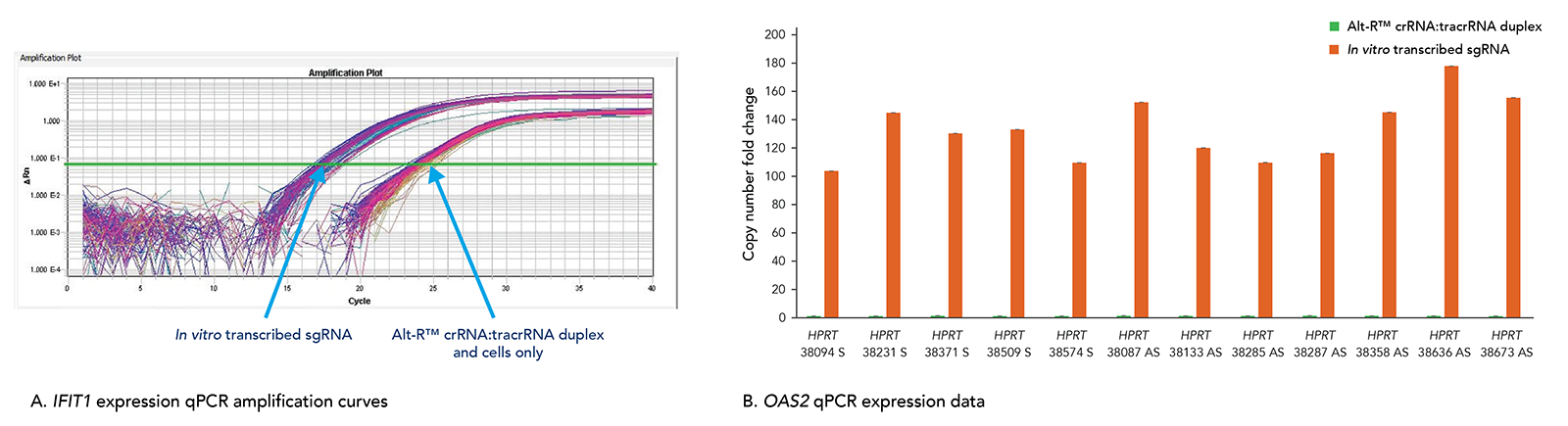
Alt-R CRISPR-Cas9 RNAs elicit less toxicity and lower innate immune response compared to in vitro transcribed guide RNA alternatives
Transfection of long in vitro transcribed (IVT) RNAs has been shown to elicit an innate immune response in our laboratories. This response can result in high cell death due to cytotoxicity. Our use of a particularly robust HEK-293–Cas9 cell line that constitutively expresses Cas9 has allowed us to compare cellular toxicity and immune response activation (Figure 6) by Alt-R RNAs and IVT RNAs. We observed high levels of activation of stress response genes such as IFIT1 (P56) and OAS2 (as well as IFITM1, RIGI, and OAS1; not shown) related to the innate immune response in cells challenged with IVT RNA triggers. These genes were not activated in cells transfected with Alt-R CRISPR-Cas9 RNAs.
Figure 6. The Alt-R CRISPR-Cas9 System does not trigger a cellular immune response. Alt-R CRISPR-Cas9 RNAs and corresponding in vitro transcribed (IVT) RNAs (triphosphate removed) designed to 12 HPRT1 sites were reverse transfected into HEK-293–Cas9 cells that stably express S.
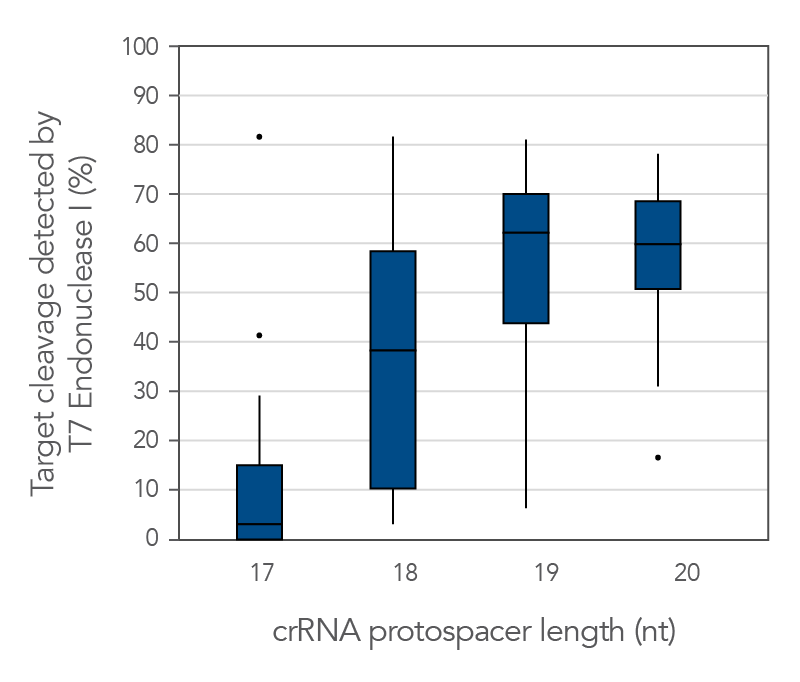
Protospacer element size is critical for editing efficacy
There are reports in the literature suggesting that CRISPR-Cas9 nuclease specificity can be improved by using truncated guide RNAs [2]. For example, 17-base protospacer elements have been reported to reduce off-target effects. We investigated how shortening protospacer element length would affect CRISPR-Cas9 nuclease specificity (Figure 7). crRNAs with protospacer element lengths of 17–20 bases were designed to 12 distinct HPRT target sites and genome editing efficiency measured using a T7EI cleavage assay (Alt-R Genome Editing Detection Kit). 20-base protospacer elements were optimal, with 19 bases providing similar strong editing efficacy in most cases. Editing efficiency was greatly reduced when 17- and 18-base protospacers were used.
Figure 7. 19–20
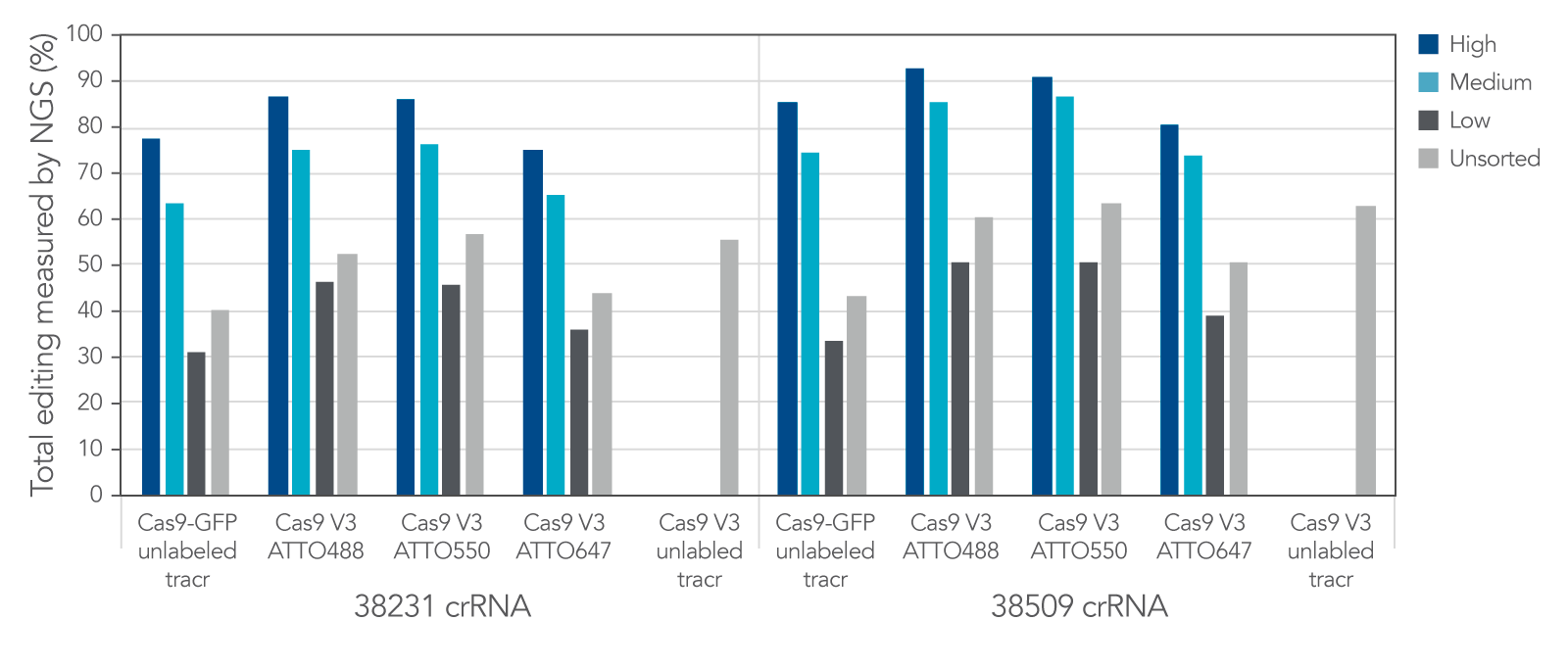
Labeled CRISPR products enable enrichment of edited cells by fluorescence signal.
IDT Cas9-GFP and fluorescent-labeled CRISPR-Cas9 tracrRNAs offer a variety of options that allow for enrichment of edited cells by fluorescence signal.
Figure 8. IDT Cas9-GFP and fluorescent labeled CRISPR-Cas9 tracrRNAs offer a variety of options that allow for enrichment of edited cells by fluorescence signal. Alt-R crRNA XT was duplexed with either unlabeled or ATTO-labeled tracrRNA and complexed with either Cas9-GFP or Alt-R Cas9 V3 and delivered into HEK293 cells using Lipofectamine RNAiMAX at 10 nM RNP. After ~18 hrs, cells were sorted using a Becton Dickinson Aria II™ cell sorter into three subpopulations, GFP High: top 20%, medium: 60–80%, and low: bottom 60% of cells, based on GFP signal. Cells were then re-plated and genomic DNA was isolated after 48–72 hrs. Editing was analyzed by NGS, n = 1 run/sample.
Cas9 nuclease
For additional data focused on Cas9 nucleases, visit the Alt-R CRISPR enzymes page.
Resources
Frequently asked questions
CRISPR genome editing efficiency seems low - any steps to improve this?
In addition, not every sequence associated with a PAM site performs the same. For example, polymorphisms in the protospacer binding site may reduce editing efficiency. Base mismatches also become more detrimental to editing the closer they are to the PAM site.
We recommend that you try 2 or 3 different PAM sites in your gene of interest to identify a site that provides optimal editing efficiency. Also, be sure to include control experiments. Alt-R™ CRISPR-Cas9 HPRT Positive Controls and Alt-R™ CRISPR-Cas9 Negative Controls are available for human, mouse, and rat.
Please feel free to contact us if you need additional assistance; having the results of your control experiments available will facilitate our ability to help you. For more information, go to www.idtdna.com/ContactUs.
When should I consider using the glycerol-free Cas9 nuclease instead of the nucleases that include glycerol?
Alt-R™ S.p. Cas9 V3, glycerol-free may be of interest when working with samples or systems where the presence of glycerol may interfere, such as primary cell cultures or high-throughput instruments with sensitive fluidics.
Do you offer CRISPR crRNA libraries?
Yes, we offer predesigned Cas9 CRISPR crRNA and additional options for creating customizable gRNA libraries.
For more information, please visit our Custom guide RNA Libraries page.
Can I use a CRISPR-Cas9 crRNA protospacer sequence that is shorter than 20 nt?
The Alt-R™ CRISPR-Cas9 crRNA ordering tool (accessible here) accommodates 19 and 20 nucleotide (nt) protospacer sequences; however, we recommend 20 nt sequences for most experiments. Other formats can be ordered as custom RNAs.
There are reports in the literature suggesting that CRISPR-Cas9 nuclease specificity can be improved through use of truncated guide RNAs [1]. For example, 17 nt protospacer elements have been reported to reduce off-target effects.
In contrast, our research investigating the effect of shorter protospacer element length on CRISPR-Cas9 nuclease specificity demonstrated that 20 nt protospacer elements were optimal, with 19 nt protospacers providing similar strong editing efficacy in most cases. When using Alt-R S.p. HiFi Cas9 Nuclease, 20 nt protospacer sequences provide the greatest amount of genomic editing.
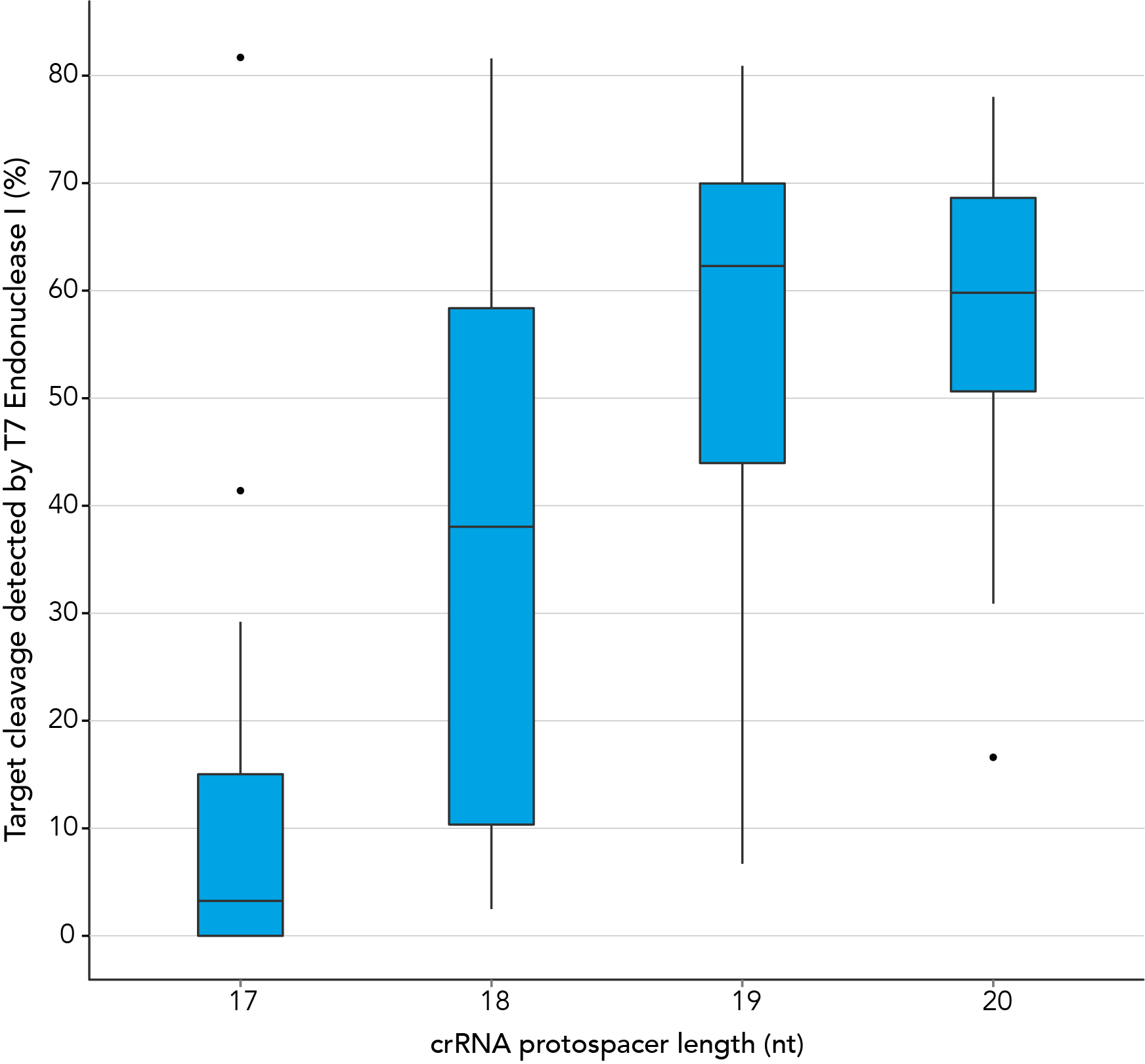 Figure 1. 19-20 nt protospacer element provides optimal genome editing. crRNAs with varying protospacer element lengths (17–20 nt) were designed to 12 distinct HPRT target sites. crRNA:tracrRNA complexes were reverse transfected into a HEK-293 cell line stably expressing S. pyogenes Cas9, using Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Genomic DNA was isolated and editing was measured by PCR amplification of target sites followed by cleavage with T7EI mismatch endonuclease (Alt-R™ Genome Editing Detection Kit) and analysis using the Fragment Analyzer™ (Advanced Analytical). At all but one of the 12 target sites, crRNAs with 19 and 20 base protospacer elements produced the greatest amount of genomic editing. Each of the 12 data points for each category represent the average of 3 biological replicates, with the exception of one data point in the 19 base category that is composed of 2 biological replicates.
Figure 1. 19-20 nt protospacer element provides optimal genome editing. crRNAs with varying protospacer element lengths (17–20 nt) were designed to 12 distinct HPRT target sites. crRNA:tracrRNA complexes were reverse transfected into a HEK-293 cell line stably expressing S. pyogenes Cas9, using Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Genomic DNA was isolated and editing was measured by PCR amplification of target sites followed by cleavage with T7EI mismatch endonuclease (Alt-R™ Genome Editing Detection Kit) and analysis using the Fragment Analyzer™ (Advanced Analytical). At all but one of the 12 target sites, crRNAs with 19 and 20 base protospacer elements produced the greatest amount of genomic editing. Each of the 12 data points for each category represent the average of 3 biological replicates, with the exception of one data point in the 19 base category that is composed of 2 biological replicates.- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279-284.


 Processing
Processing