SARS-CoV-2 coronavirus sequencing
Targeted NGS is a valuable research tool for the detection and characterization of viral genomes. We offer solutions designed for SARS-CoV-2 variant research.
Discovery
Flexibility
Speed
Overview
Next generation sequencing (NGS) can provide valuable information about the spread and genetic evolution of a virus. Targeted NGS is a valuable basic research tool for both identification and characterization of viruses. Since targeted NGS research provides data about viral genetic changes, the data can be used in combination with basic disease research to potentially correlate genetic changes that increase transmissibility or severity of disease.
Why sequence the SARS-CoV-2 genome?
The genome sequences we get from sequencing SARS-CoV-2 are crucial to track and trace the ongoing outbreak of COVID-19. Using NGS, we can look at the virus’ genomic sequence from virus-positive samples to help us understand where it may have originated, how it is mutating or evolving, and where it spreads. Knowing the genome sequence information is also critical to research:
- Origin identification
- Track mutations and evolution rates
- Identify transmission routes
- Population surveillance
Methods for NGS viral sequencing
There are several methods that can be used by researchers to study the SARS-CoV-2 viral genome:
- Whole genome sequencing (WGS)
- Target enrichment via multiplex PCR (amplicon sequencing)
- Target enrichment via hybridization capture
Whole genome sequencing is a rapid way of getting from sample to sequencing and can be a good method to utilize if one is looking for a comprehensive picture of all pathogens in a sample. However, if the interest is solely to study SARS‑CoV‑2, it’s important to keep in mind that samples will also contain host or human genetic material, which could make sequencing inefficient and potentially increase associated costs.
Target enrichment methods are an efficient and cost-conscious way to study the complete SARS-CoV-2 viral sequence, or even select genes from its genome. These methods enrich for those desired regions prior to library prep or sequencing.
IDT target enrichment optoins:
Target enrichment via multiplex PCR (amplicon sequencing):
Following RNA isolation, this method requires relatively few steps which makes it faster to get from sample to sequencing without sacrificing genome coverage.
xGen SARS-CoV-2 Amplicon Panel (formerly the Swift Normalase™ Amplicon SARS-CoV-2 Panel—SNAP SARS Panel)
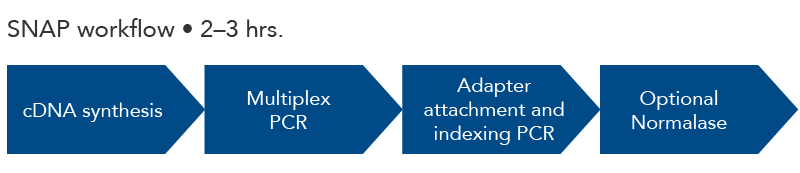
One-tube amplicon sequencing library preparation in 2-3 hours time. The optional Normalase step creates equimolar amounts of each library for multiplexing. The final library is compatible with Illumina sequencing instruments for understanding the changing sequences of viral isolates.
Figure 1. xGen SARS-CoV-2 Amplicon Panel (formerly Swift SNAP-SARS Panel). The steps for amplicon sequencing with the xGen SARS-CoV-2 Amplicon Panel include cDNA synthesis (reagents not included), amplicon generation with multiplex PCR, adapter ligation/indexing by PCR, and a final optional normalization step to create equimolar amounts of multiplexed library samples.
xGen SARS-CoV-2 ARTIC Amplicon Panel
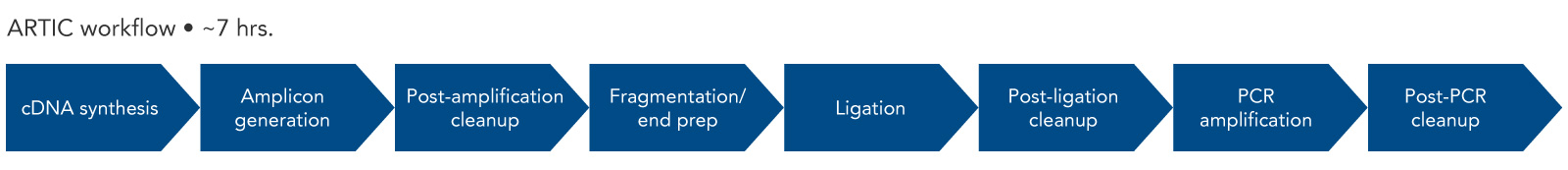
Amplicons are 400 bp long, making them compatible with either Oxford Nanopore® or Illumina® sequencing instruments.
Figure 2. ARTIC workflow. The steps for amplicon sequencing with the ARTIC Panel and the IDT DNA Library Prep Kit include cDNA synthesis, amplicon generation, fragmentation, adapter ligation, and a final PCR amplification. The combination results in Illumina or Nanopore-ready libraries.
xGen SARS-CoV-2 Midnight Amplicon Panel
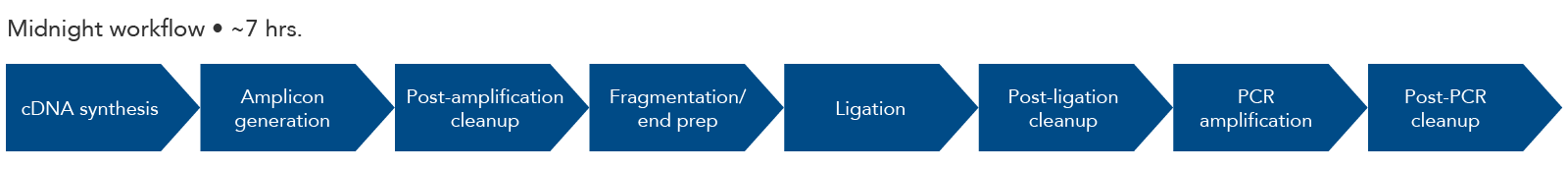
Amplicons are 1200 bp long, making them compatible with the Oxford Nanopore long-read sequencing system. To make these amplicons compatible with Illumina sequencing instruments, use one of IDT's xGen DNA Library Prep Kits to create smaller library fragments.
Figure 3. Midnight panel workflow. The steps for amplicon sequencing with the Midnight Panel and the IDT Lotus DNA Library Prep Kit include cDNA synthesis, amplicon generation, fragmentation, end prep, adapter ligation, and a final PCR amplification. The final library is ready for sequencing with Illumina sequencing instruments.
Target enrichment via hybridization capture:
Another targeted next generation sequencing method that uses long, biotinylated oligonucleotide baits (probes) to hybridize to regions of interest. It is particularly helpful for enriching viral genomes when sequencing a sample.
xGen SARS-CoV-2 Hyb Panel
Available as a bundle with xGen Universal Blockers-TS Mix and xGen Hybridization and Wash Kit. For more information, see the xGen SARS-CoV-2 Hybridization Panel.
NYU Grossman School of Medicine
ALIGN Program: SARS‑CoV‑2 sequencing service providers
IDT partners with some of the most comprehensive genomic sequencing providers in the world. These companies are aligned in our commitment to collaboration, and we are aligned in our resolve to break down research barriers. Whether you’re focused on whole genome sequencing or custom panels, our partners have you covered.
More about the ALIGN programResources
Press releases
References
- Freed NE, Vlkova M, Faisal MB, et al. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding. Biol Methods Protoc. 2020;5(1):bpaa014.


 Processing
Processing