Mutant and modified CRISPR-Cas enzymes
Overview
Mutants and fusion proteins of Cas enzymes have been produced to make nickases, catalytically inactive (dead) Cas enzymes, base editors, many functional fusion proteins, and a “prime editing” enzyme. These modified enzymes have a wide range of functions.
Mutant CRISPR-Cas enzymes
CRISPR is such a new technology that it is hard to say that certain approaches are “classical,” while others are newer. However, if creating double-strand breaks (DSBs) in DNA is considered the “classical" approach to CRISPR genome editing, you can compare the creation of DSBs to some other approaches that scientists have taken by making a wide variety of Cas9 mutants and fusion proteins.
The CRISPR basics handbook
Everything you need to know about CRISPR, from A to Z, from theory to practice, for beginners as well as advanced users.
Cas9 nickases and catalytically dead Cas9
Native Cas9 enzyme naturally makes DSBs in DNA. The enzyme has two protein domains, HNH and RuvC, each of which cuts a different strand of the DNA. The HNH domain cleaves the targeted DNA strand, while the RuvC domain cleaves the non-targeted DNA strand, which contains the PAM sequence. By mutating one of the two protein domains, scientists have made “nickase” forms of Cas9. Cas9 nickase variants have an alanine substitution within one of these catalytic domains that renders the mutated domain inactive. Therefore, Cas9 nickases cut one strand of DNA without cutting the other strand, creating a nick. In the RuvC mutant (D10A) nickase, only the HNH domain is active, so the cut is on the targeted strand. Conversely, H840A nickase, the HNH mutant with active RuvC, cuts the non-targeted strand. These nickases are both available from IDT.
By mutating both the HNH and RuvC domains of Cas9, scientists have been able to block the DNA-cleaving activity of Cas9. This catalytically inactive form of Cas9 is known as dCas9, which stands for “dead Cas9.” This variant still binds to targeted DNA when directed there by a guide RNA, but it does not cleave the DNA. However, it still prevents other enzymes such as transcription activators from binding that site. Thus, transcription is blocked, and the gene is temporarily silenced [1]. When dCas9 is naturally cleared by the cell, transcription can resume. In this convenient way, dCas9 can be used for reversible gene-silencing experiments. Recombinant dCas9 is available from IDT.
Base editor systems and other Cas9 fusion proteins
Scientists have fused Cas9—both catalytically active and inactive—with many other proteins. Base editor systems are one notable example of the use of fusion proteins. In these systems, dCas9 is fused with a nucleobase deaminase enzyme [2]. These fusion proteins precisely target a site in DNA and directly exchange one base for another. Such enzymes are good at making single point mutations at desired locations without depending on cellular machinery to repair DSBs, so they are particularly useful in some non-dividing cells.
Other examples of functional Cas9 fusion proteins include Cas9 fused with enhanced green fluorescent protein (eGFP) or light-responsive cryptochrome [4,5].
CRISPR prime editing
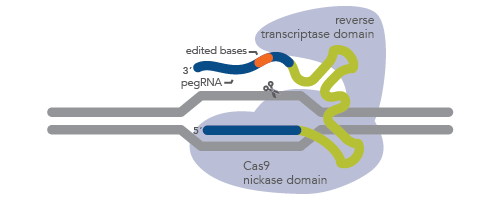
One promising approach to CRISPR is known as CRISPR prime editing (Figure 1). In this approach, a Cas9 nickase is fused with an engineered reverse transcriptase enzyme. What makes the prime editing approach unique is the use of a prime editing guide RNA (pegRNA). This pegRNA is a long, single-guide RNA with extra bases that serve as a template for synthesis of a new DNA sequence [3]. pegRNA directs the enzyme to the target site as in normal CRISPR genome editing but also loops across the complex to the other side of the DNA to dictate the new sequence. For more information, see the article, Prime editing with pegRNA. IDT manufactures pegRNA for CRISPR prime editing.
Figure 1. Schematic representation of pegRNA in CRISPR prime editing. Prime editing uses a Cas9 H840A nickase–reverse transcriptase fusion protein (light blue) and a long guide RNA called pegRNA. pegRNA is composed of targeting RNA (the lower dark blue line), an enzyme-binding region (green line), and a region pairing to the cut strand of DNA (the upper dark blue line). The orange region represents the new (edited) sequence. Genomic DNA is depicted as gray lines.
Other Cas enzymes
Among the many Cas enzymes, several have been studied in the context of genome editing. Cas12a is another widely popular Cas used for “classical” (DSB-forming) genome editing and is available as recombinant protein from IDT. In some labs, Cas12a, like Cas9, has also been rendered catalytically dead and/or fused to other proteins to enable alternative uses [6].
Application note: optimized method for CRISPR-Cas9 HDR experiments
This application note explores the optimization of HDR in mammalian cell lines, including criteria for selecting guide RNA sequences, considerations for DNA donor template design, as well as use of either Cas9 nuclease or Cas9 nickase.
Related Products
Alt-R™ CRISPR nucleases
Recombinant, high-purity Cas9 and Cas12a (Cpf1) endonucleases for genome editing experiments.
Alt-R™ CRISPR Custom Guide RNAs
Custom guide RNAs ideal for prime editing (pegRNA) projects, CRISPR-Cas13 applications, and most alternative CRISPR-Cas systems.
References
- Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression [published correction appears in Cell. 2021 Feb 4;184(3):844]. Cell. 2013;152(5):1173-1183.
- Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells [published correction appears in Nat Rev Genet. 2018 Oct 19;:]. Nat Rev Genet. 2018;19(12):770-788.
- Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149-157.
- Zhou Y, Wang P, Tian F, et al. Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging. Cell Res. 2017;27(2):298-301.
- Gangopadhyay SA, Cox KJ, Manna D, et al. Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry. 2019;58(4):234-244.
- Xu X, Qi LS. A CRISPR-dCas Toolbox for Genetic Engineering and Synthetic Biology. J Mol Biol. 2019;431(1):34-47.


 Processing
Processing