miRNA Inhibitors
Steric blocking oligonucleotides hybridize with mature miRNAs (microRNAs) and inhibit their functions. IDT miRNA Inhibitors are single-stranded oligonucleotides comprised of 2’-O-methyl residues that confer increased binding affinity to RNA targets and resistance to endonuclease degradation. ZEN modifications are included to block exonuclease degradation.
Ordering
- Effectively inhibit miRNA function in vitro using low nanomolar concentrations
- Ensure sufficient miRNA knockdown with highly potent, highly specific reagents
- Simplify your experiments with intuitive design and ordering tools
Product Details
The standard method for inhibiting microRNA (miRNA) function is by steric blocking—using an oligonucleotide that is perfectly complementary to the mature miRNA target. miRNA inhibitors form a duplex with the miRNA guide strand that prevents the miRNA from binding to its intended target. miRNA function is based on recognition of a seed region, rather than complete homology between miRNA and target. A single miRNA can regulate tens to hundreds of genes whose sequences do not share exact complementarity with the miRNA.
For effective miRNA inhibition, the binding affinity between the oligo inhibitor and the miRNA must be significantly higher than that between the miRNA guide strand and the passenger strand. The miRNA inhibitor must be capable of binding to the miRNA guide strand either in single-stranded form, or when bound to an Argonaute protein in a miRNA-induced silencing complex (miRISC). The inhibitor should also be capable of displacing the natural passenger strand in double-stranded miRNA.
Adding a 2′ modification to ribose sugars in an RNA oligonucleotide can increase melting temperature (Tm) and confer resistance to nucleases [1]. 2′-O-methyl RNA (2′OMe) is a naturally occurring, nontoxic nucleic acid with a high binding affinity for RNA that provides the required resistance to mammalian endonucleases. Adding ZEN modifications to the termini of 2′OMe-modified oligonucleotides further increases the binding affinity of the miRNA inhibitors and gives resistance to exonucleases [1].
Where to find miRNA sequences
Mature miRNA sequences can be found in the miRNA database, miRBase [2–5]. Copy the mature sequence for each miRNA, then paste it into the IDT miRNA Inhibitor ordering tool.
Controls
We recommend using the following positive and negative controls for your miRNA modulation experiments.
Positive controls
- miRNAs are expressed at various levels in different cell types so it can be tricky to find a good positive control. Make sure the positive control that you select is expressed at sufficiently high levels to provide an adequate measurement of the response.
- A miRNA that serves as a good positive control in many situations is miR-21-5p; it is conserved in many species and expressed at high levels in many cell types. Protein modulation can be confirmed by measuring the expression of endogenous miR-21 targets, such as PTEN [6] and PDCD4 [6–7], or using a reporter assay.
| Species | Mature miR-21-5p sequence* (copy and paste into IDT miRNA Inhibitor ordering tool) |
|---|---|
| Human, mouse, rat | uagcuuaucagacugauguuga |
*Note: The IDT miRNA ordering tool will convert the mature miRNA sequence to the complementary sequence with 2′OMe and ZEN modifications added for stability. Mouse miRNA is miR-21a-5p.
Negative controls
- A good negative control should be inert and not modulate any genes in the system under study. Since this is hard to achieve, we propose two negative control sequences that we have used throughout our product development and validation process which were found to work well with both in cellulo and in vivo.
| Species | Sequence† (copy and paste into IDT miRNA Inhibitor ordering tool) |
|---|---|
| NC1 Negative control (human) | ucguuaaucggcuauaauacgc |
| NC5 Negative control (human, mouse, rat) | accauauugcgcguauagucgc |
†Note: The IDT miRNA ordering tool will convert the mature miRNA sequence to the complementary sequence with 2′OMe and ZEN modifications for added stability.
Product Data
The success of miRNA inhibition experiments is dependent on a number of factors—most importantly: potency, selectivity, stability, and safety. Higher potency requires lower doses to produce the desired phenotype while also reducing toxicity of the administered compound. Those miRNA inhibitors that are specific to their target reduce the incidence of off-target effects, ensuring that any observed phenotypes are the result of the effect on the target under investigation. Stability and low toxicity are essential for in vivo experiments.
Figures 1–4 demonstrate the high potency, stability, and low toxicity of IDT miRNA Inhibitors compared to other modified oligonucleotides.
Modifications tested in miRNA inhibition experiments
Chemically modified oligonucleotides were designed to target miR-21. The sequences are the same, but their modifications differ. See Table 1 and Figures 1–4 for experimental results.
Table 1. Varying chemical modifications of the same oligonucleotide.
| Name | Sequence‡ |
|---|---|
| IDT miRNA Inhibitors | mU/ZEN/mCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmUmA/3ZEN/ |
| DNA | TCAACATCAGTCTGATAAGCTA |
| 2′OMe | mUmCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmUmA |
| 2′OMe 3PS Ends | mU*mC*mA*mAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmG*mC*mU*mA |
| 2′OMe 3′inZEN | mUmCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmU/ZEN/mA |
| 2′OMe 5′inZEN | mU/ZEN/mCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmUmA |
| 2′OMe 3′inC3 | mUmCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmU/iSpC3/mA |
| 2′OMe 5′inC3 | mU/iSpC3/mCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmUmA |
| 2′OMe 5′inC3, 3′inC3 (2′OMe 2XC3) | mU/iSpC3/mCmAmAmCmAmUmCmAmGmUmCmUmGmAmUmAmAmGmCmU/iSpC3/mA |
‡A/T/C/G = DNA base; mA/mU/mC/mG = 2′ O-methyl (2′OMe) RNA base; * = phosphorothioate (PS) linkage; /ZEN/ or /3ZEN/ = internal or 3′ ZEN modification, respectively; /iSpC3/ = internal C3 spacer.
High potency of IDT miRNA Inhibitors
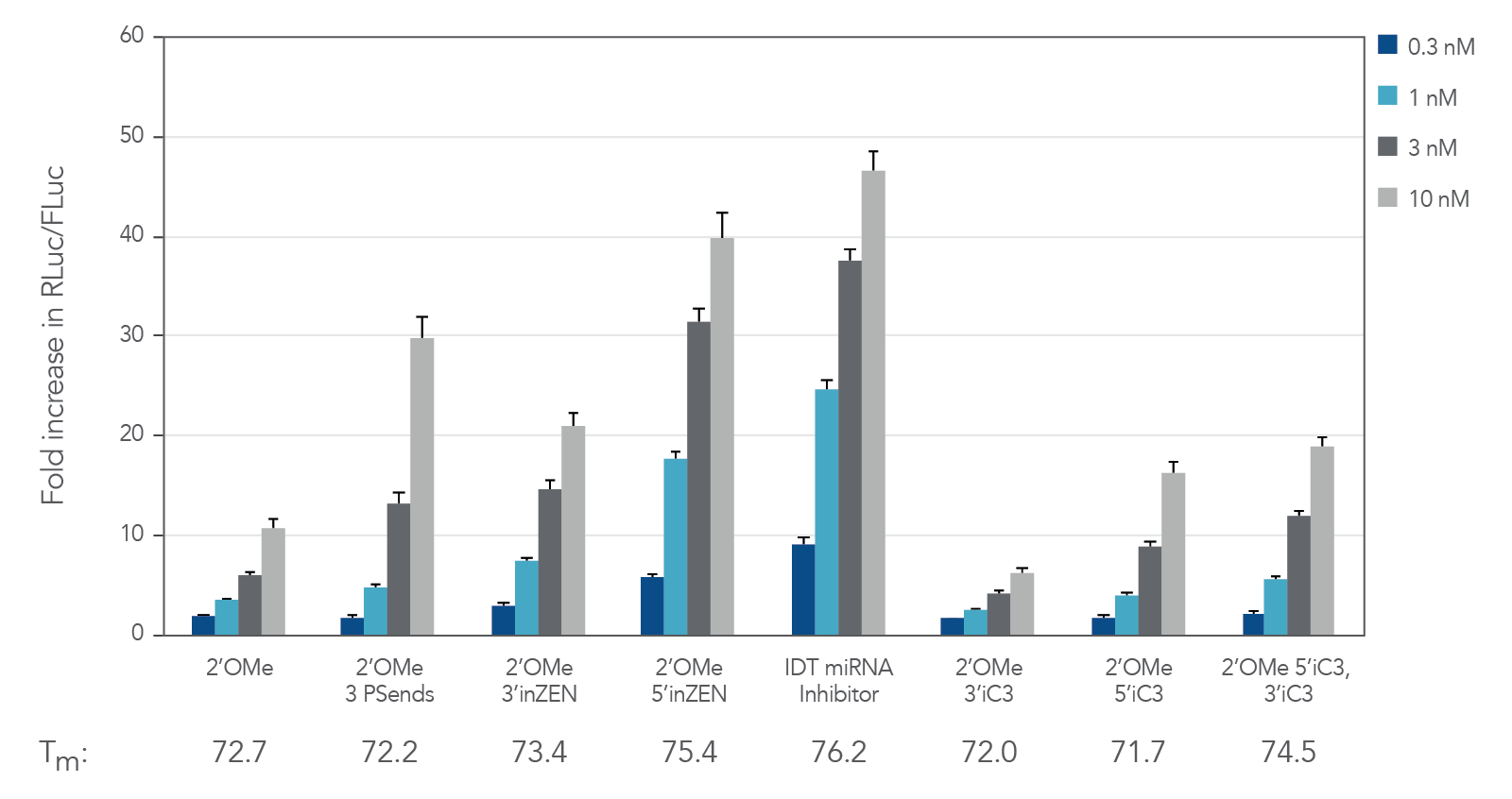
Figure 1. IDT miRNA Inhibitors exhibit high potency. Oligonucleotides designed to target miR-21 were transfected at 0.3–30 nM in HeLa cells expressing the psiCHECK-miR-21 plasmid using Lipofectamine® RNAiMAX transfection reagent (Thermo Fisher Scientific). The cells were lysed after 24 hr and analyzed for luciferase activity. Results were normalized with the internal firefly luciferase (FLuc) control and are shown as fold change in Renilla luciferase ( RLuc) compared with the lipid reagent control, which was set at 1. Below their respective profiles, Tm values for the various oligos are shown. Error bars represent the standard error of the mean (SEM), n = 3.
High specificity of IDT miRNA Inhibitors
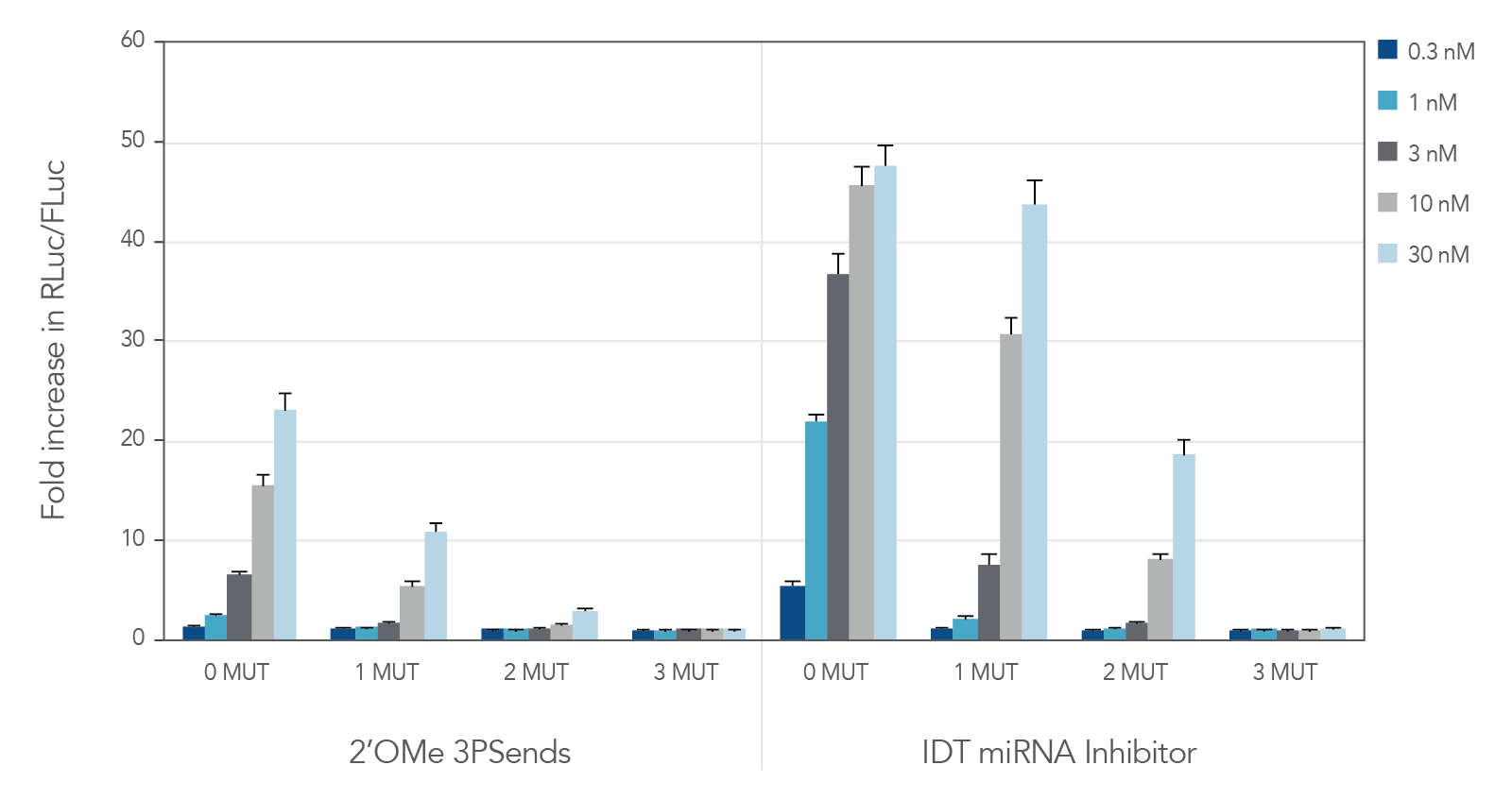
miRNA inhibitors with various modifications were tested against wild-type miR-21 and 3 "mutant" versions containing 1, 2, or 3 mismatches (Table 2). IDT miRNA Inhibitors demonstrated high selectivity, especially when ≥ 2 mismatches are present (Figure 2).
Table 2. Sequences of wild-type and "mutant" miR-21 used to test specificity of miRNA inhibitors.
| Mutant type | miR-21 IDT miRNA Inhibitor sequences (5′ to 3′)§ |
|---|---|
| Wild type (0 MUT) | C A A C A U C A G U C U G A U A A G C U |
| 1 MUT | C A A C A U C A G U C A G A U A A G C U |
| 2 MUT | C A A C C U C A G U C A G A U A A G C U |
| 3 MUT | C A A C C U C A G U C A G A U A A C C U |
§Mutated bases are indicated by bold, red notation. Modifications not shown.
Figure 2. IDT miRNA Inhibitors are highly specific. Different miR-21 inhibitors (n = 5) were synthesized, complementary to wild-type miR-21 (0 MUT), or containing 1, 2, or 3 mismatches—1 MUT, 2 MUT, and 3 MUT, respectively. The inhibitors were transfected into HeLa cells expressing the psiCHECK-miR-21 plasmid. After 24 hr, the cells were lysed and analyzed for luciferase activity. Values were normalized with internal firefly luciferase (FLuc) control and reported as a fold change in Renilla luciferase (RLuc), compared with lipid reagent control set at 1. Error bars represent the standard error of the mean (SEM), n = 3.
High nuclease stability of IDT miRNA Inhibitors
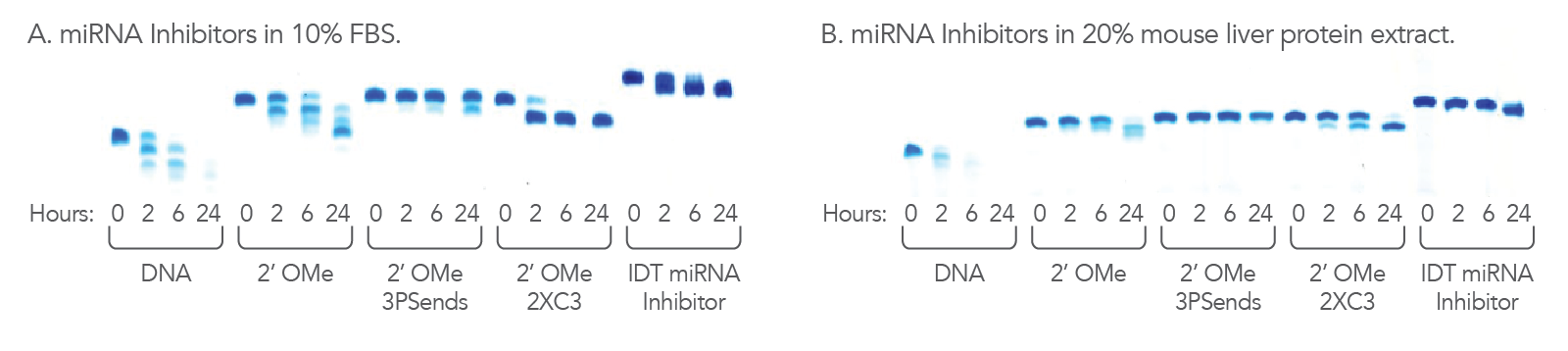
Figure 3. IDT miRNA Inhibitors are resistant to nucleases. 1.1 nmol of each oligonucleotide was incubated in (A) 10% FBS, high exonuclease environment; or (B) 20% mouse liver cell extract, an exo- and endonuclease containing environment, for the indicated lengths of time. Each reaction was analyzed on a denaturing polyacrylamide gel stained with methylene blue.
Low cellular toxicity of IDT miRNA Inhibitors
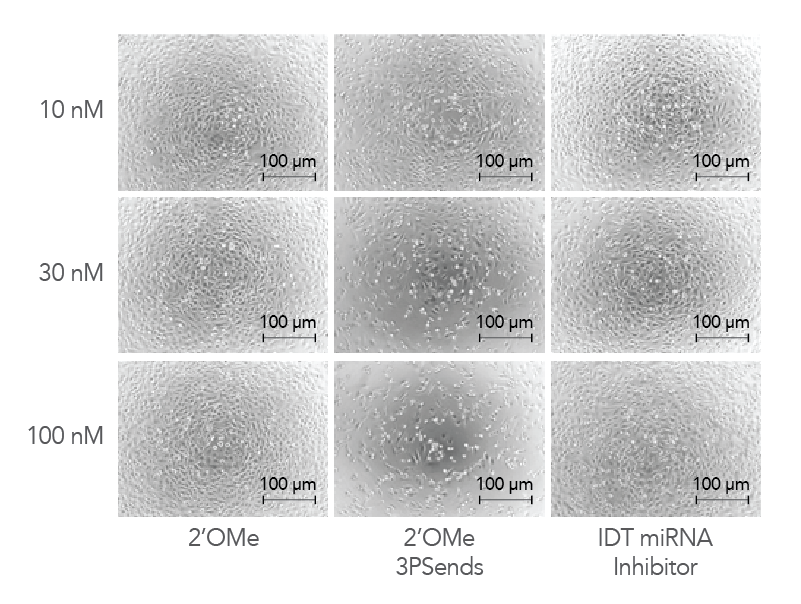
Figure 4. IDT miRNA Inhibitors exhibit low toxicity to cells. Modified oligonucleotides corresponding to a non-targeting negative control sequence, NC1, were transfected into HeLa cells at concentrations of 10, 30, and 100 nM to investigate and compare toxicity. The cells were visualized by phase contrast microscopy (10X magnification).
GMP∥ and OEM
If you require oligos that are approved for use in molecular diagnostics applications, or if you are interested in our OEM services, please click here.
∥GMP refers to products manufactured under ISO 13485: 2016 QMS. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations for their legal marketing.
Resources
Frequently asked questions
What are miRNA inhibitors (AMOs) and how are they used?
Anti-microRNA Oligonucleotides (AMOs), or microRNA inhibitors, are steric blocking antisense reagents that inhibit microRNA (miRNA) function by hybridizing to and repressing the activity of a mature miRNA.
The miRNA regulatory networks exert some level of control in the majority of cellular biological processes, including cell differentiation, apoptosis, and proliferation. AMOs/microRNA inhibitors can be used to inhibit miRNA function, both to investigate that function and for therapeutic purposes to correct diseases associated with miRNA dysregulation.
You can read more about as well as order IDT miRNA Inhibitors at miRNA Inhibitor product page
References
- Lennox KA, Owczarzy R, Thomas DM, et al. Improved performance of anti-miRNA oligonucleotides using a novel non-nucleotide modifier. Mol Ther Nucleic Acids. 2013;2(8):e117.
- Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152-157.
- Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154-158.
- Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140-144.
- Frankel LB, Christoffersen NR, Jacobsen A, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026-1033.
- Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27(31):4373-4379.
- Yao Q, Xu H, Zhang QQ, et al. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388(3):539-542.


 Processing
Processing